Sugar uptake
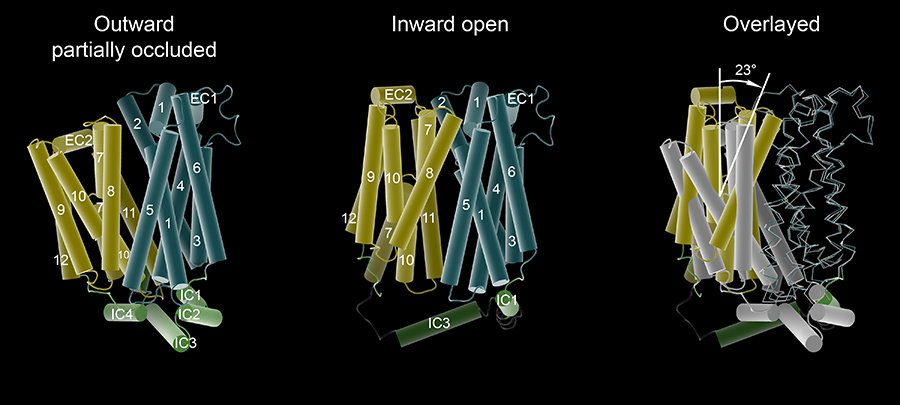
The major facilitator superfamily of membrane proteins is the largest collection of structurally related membrane proteins that transport a wide array of substrates. The proton-coupled sugar transporter XylE is the first member of the MFS that has been captured and structurally characterized in multiple transporting conformations including both the outward and inward facing states. We determined the crystal structure of XylE in a new inward-facing open conformation. Structural comparison of XylE in this conformation with its outward-facing partially occluded conformation reveals how this transporter functions through a non-symmetrical rocker switch movement of the N-domain as a rigid body and the C-domain as a flexible body. Molecular dynamics simulations were employed to help describe how XylE transitions in a lipid membrane to facilitate sugar transport.
Nitrogen uptake
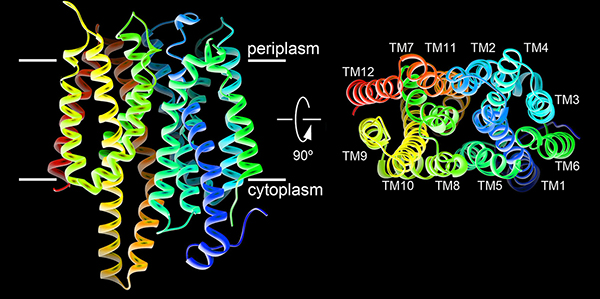
Nitrate is the preferred nitrogen source for plants on which all higher forms of life ultimately depend. Plants and microorganisms evolved to efficiently assimilate nitrate. Despite decades of effort no structure was available for any nitrate transport protein and the mechanism by which nitrate is transported remained largely obscure until our study was published. We reported the structure of a bacterial nitrate/nitrite transport protein, NarK, from Escherichia coli, with and without substrate. The structures revealed a positively charged substrate-translocation pathway lacking protonatable residues, suggesting that NarK functions as a nitrate/nitrite exchanger and that H⁺s are unlikely to be co-transported. Conserved arginine residues form the substrate-binding pocket, which is formed by association of helices from the two halves of NarK. Key residues that are important for substrate recognition and transport were identified and related to extensive mutagenesis and functional studies. We proposed that NarK exchanges nitrate for nitrite by a rocker-switch mechanism facilitated by inter-domain H-bond networks.
Amino acid uptake
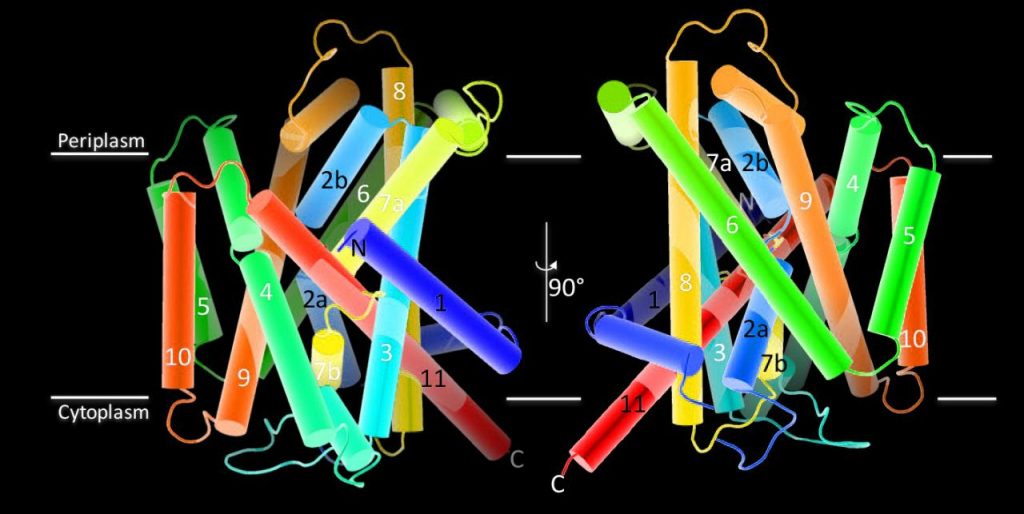
The amino acid, polyamine, and organocation (APC) superfamily is the second largest superfamily of membrane proteins forming secondary transporters that move a range of organic molecules across the cell membrane and that can transport both D- and L-amino acids. Here we report two crystal structures of an APC member from Methanococcus maripaludis, the alanine or glycine:cation symporter (AgcS), with L- and D-alanine. Structural analysis combined with site-directed mutagenesis and functional studies inform on substrate binding, specificity, and modulation of the AgcS family and reveal key structural features that allow this transporter to accommodate glycine and both L- and D-type alanine while excluding all other amino acids. Mutation of key residues in the substrate binding site expand the transporters selectivity to include valine and leucine. Moreover, as a transporter that binds both enantiomers of alanine, the present structures provide an unprecedented opportunity to gain insights into the mechanism of stereo-selectivity in membrane transporters.
Relevant papers
Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE
In: Nat Commun, vol. 5, pp. 4521, 2014.
Crystal structure of a nitrate/nitrite exchanger
In: Nature, vol. 497, no. 7451, pp. 647–651, 2013.
In: Structure, vol. 18, no. 11, pp. 1512–1521, 2010.
The prototypical H⁺/Galactose Symporter GalP Assembles into Functional Trimers
In: J. Mol. Biol., vol. 396, no. 3, pp. 593–601, 2010.